
The MEAP-IHNV database contains over 1000 records that provide background information and genetic sequencing data for individual isolates of the fish virus Infectious hematopoietic necrosis virus (IHNV). The database focuses on IHNV isolates collected throughout Western North America from 1966 to the present. It also includes a smaller number of IHNV isolates from Eastern Russia, and in the future we will add a small number of representative IHNV isolates from Europe and Asia. The purpose of this database is to provide access to detailed data for all parties interested in IHNV molecular epidemiology, such as fish health professionals, fish culture facility managers, and academic researchers. The database has flexible search capabilities and generates various output formats, including tables and maps, that we hope will assist users in developing and testing hypotheses about how IHNV moves across aquatic landscapes and changes over time. By engaging the expertise of the broader community of colleagues interested in IHNV, our goal is to enhance the overall understanding of IHNV epidemiology, including defining sources of disease outbreaks and viral emergence events, identifying virus traffic patterns and potential reservoirs, and understanding how human management of salmonid fish culture impacts disease. Ultimately this knowledge will be used to develop new strategies to reduce the impact of IHN disease in cultured and wild fish.
IHN virus is a well known pathogen of salmonid fish, with a host range including many species of salmon and trout. Taxonomically it is the type species of the genus Novirhabdovirus, within the virus family Rhabdoviridae, which includes other well known animal viruses such as rabies virus and vesicular stomatitis virus. IHNV originated in western North America, where it is currently endemic to most Pacific watersheds that contain salmonid fish. In the 1970s and 1980s IHNV was introduced to Europe and Asia by transport of virus-contaminated fish eggs. IHNV can be transmitted either horizontally through waterborne virus, or vertically as egg-associated virus. Infection can result in lethal necrosis of the hematopoietic tissues of the kidney and spleen, and disease outbreaks can cause up to 90% mortality depending on various host, virus, and environmental factors. IHNV is a significant impediment to rainbow trout farm culture, and to hatchery and netpen programs that rear salmon and trout. Due to the economic impact of IHNV, spawning adults and juvenile fish in most cultured salmonid populations in western North America are routinely surveyed for IHNV.
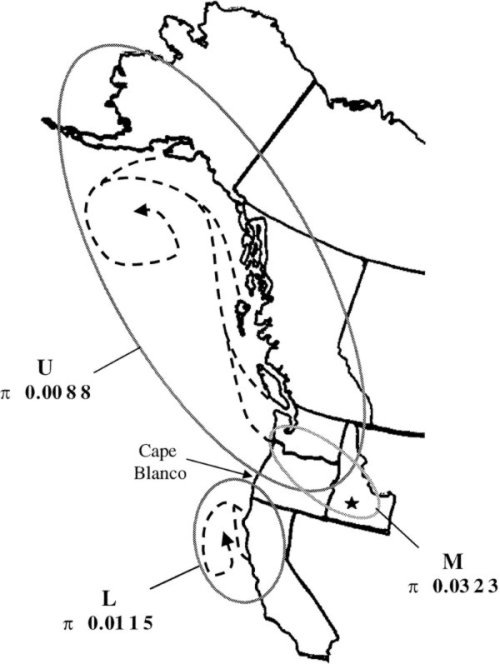
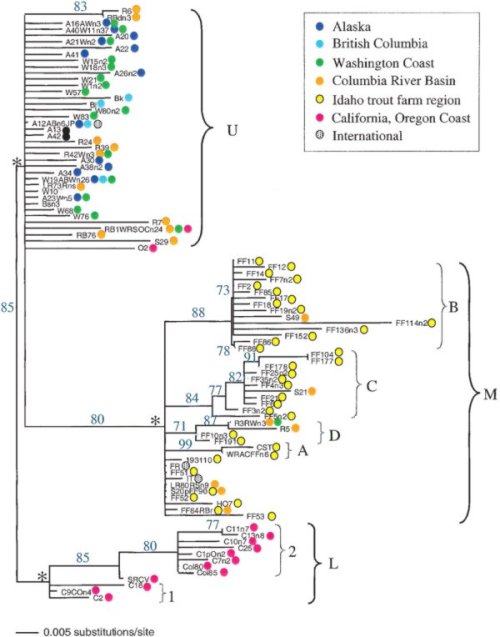
Several molecular epidemiology studies of North American IHNV field isolates have revealed three major genogroups of IHNV, designated U, M, and L because they occur in the upper, middle, and lower portions of the IHNV geographic range in North America (Figures 1 and 2). Although each genogroup can infect multiple fish host species, the U, M, and L groups show some host specificity for sockeye salmon (Oncorhynchus nerka), rainbow and steelhead trout (O. mykiss), and Chinook salmon (O. tshawytscha), respectively.
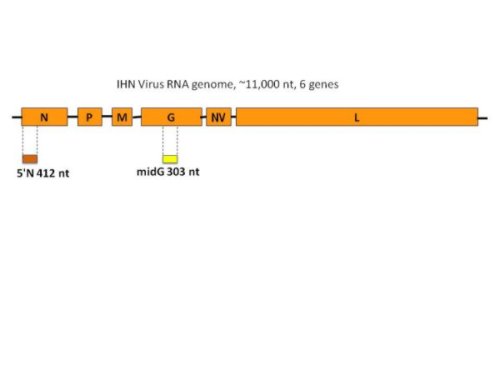
IHNV has a single-stranded, negative sense RNA genome of approximately 11,000 bases. This genome has six genes, which are ordered along the genome as 3' N (nucleocapsid), M (matrix), P (phosphoprotein), G (glycoprotein), NV (non-virion), and L (large polymerase)-5'. The G gene is approximately 1610 nucleotides (nt) long and encodes the only protein on the outer surface of the enveloped virus particles. Standard genetic typing was done by sequencing a variable 303 nt region in middle of the G gene, referred to as the "midG". Where additional information was desired, a 412 nt partial gene sequence at the 5' end of the N gene, known as the "5'N", has been sequenced for some isolates, and complete G or N gene sequences have been included where available. These sequence regions are shown in Figure 3. The midG sequence has been determined for all isolates in the database. Each different midG sequence is referred to as a "genotype", and any virus isolates with that exact same midG sequence that would be grouped within that genotype. Thus, some genotypes represent single isolates, and others may represent a large number of isolates. A universal nomenclature system has been developed to describe these genotypes. The "Universal Sequence Designator", or USD, for midG genotypes is written as "mGXXXg", where XXX is a randomly assigned 3 digit number identifying a specific midG sequence, and g indicates the major genogroup U, M, or L. For example, mG001U is the USD for a common midG sequence (genotype) in the U genogroup, and mG007M is a common genotype in the M genogroup. The 3 digit numbers are only used once in the system, regardless of which genogroup the sequence falls into. A similar system is used to refer to 5'N sequences, where each USD is written as 5NXXXg.
The database can be searched by an actual genetic sequence, or by using the USD designation, to identify all isolates with identical genotypes. The value of the USD system is that it allows us to talk about what the genetic typing data tells us about IHNV epidemiology. Isolates with the same USD are likely more closely related than isolates with different USDs, although it is important to remember that the midG region is short and additional sequence differences may well occur outside this region. To date we find that the basic resolution of the major genogroups U, M, and L, is consistent in phylogenetic trees based on midG, 5'N, or complete G sequences where they are available. Isolates with the same USD may indicate epidemiologically linked isolates, and they should be considered for spatial, temporal, or host links. When isolates have different USDs the sequence differences function as tags or markers that can be useful for tracking the virus over landscapes or through time. Single nt differences suggest close relationships, and may represent relatively recent divergence by mutation compared with isolates that have multiple nt differences. As a simple example of interpretation, if two isolates from the same collection location differ in midG sequence by 1 nt, they could have arisen by a relatively recent mutation that occurred at that location to create a variant, or by independent introductions of the two different types. However, if two isolates from the same location differ by 5 nt in the midG, that more strongly suggests two introduction events. It is important to keep in mind that without additional testing sequence differences at this level cannot be assumed to indicate any biological or phenotypic difference between isolates,
The methods used here for genetic typing of IHNV field isolates were developed in the mid-1990s in the laboratory of Gael Kurath at the U.S.G.S. Western Fisheries Research Center (WFRC) in Seattle, Washington. Virus isolates and their associated background information were obtained from numerous state, federal, and tribal fish health colleagues. A virus "isolate" is defined as the virus produced in cultured fish cells inoculated with a fish tissue sample collected from a specific fish, or pool of fish, at a specific time and location. Most sequences in this database were generated in the Kurath laboratory at WFRC. Some Oregon isolates were sequenced by John Kaufman of the Oregon Department of Fish and Wildlife, and some California isolates were sequenced in the laboratory of Ron Hedrick at the University of California, Davis. Russian IHNV isolates were sequenced in collaboration with Svetlana Rudakova of the Kamchatka Research Institute of Fishery and Oceanography.
This database does not represent all IHNV that has been isolated during fish health surveillance in North America. There are numerous state, federal, tribal, and private fish health laboratories that routinely conduct diagnostic sampling for virus in various juvenile and adult salmonid populations throughout western North America. Among the large number of virus-positive samples from these efforts, only a subset submitted for genetic typing are represented here. Typically these are selected to provide information relevant to specific epidemiological events such as disease outbreaks, or as part of local or regional studies of IHNV genetic diversity. The isolates included here do not necessarily represent consistent geographic or temporal coverage of the virus in North America. We have attempted to characterize virus from diverse geographic areas, and from hosts with diverse characteristics. However, in general our temporal coverage includes more isolates from 1990 to present, with fewer older isolates due to availability. Spatially some collection sites are represented by many isolates and some have only one or a small number, depending on the aims of the study that involved the genetic typing. The database does not have equal coverage of IHNV from wild and cultured salmonids. The database is biased toward virus isolates from cultured salmonids in hatcheries, netpens, spawning channels, and fish farms, due to the greater sampling effort expended in health care for these populations. Thus, isolates from wild fish comprise approximately 10% of the database. This database does not include IHNV isolates from Europe or Asia. Information on European IHNV can be found in the Fish Pathogens.eu database available online at http://www.fishpathogens.eu/ihnv, and both European and Asian IHNV are described in published scientific literature.
Bootland LM, Leong JC. 1999. Infectious hematopoietic necrosis virus. In: Fish Diseases and Disorders, Vol.3 (ed. by P.T.K.Woo & D.W. Bruno), pp.57-121. CAB International, New York, NY.
Emmenegger EJ, Meyer TR, Burton TO, Kurath G. 2000. Genetic diversity and epidemiology of infectious hematopoietic necrosis virus in Alaska. Dis. Aquat. Org. 40:163-176.
Emmenegger EJ, Kurath G. 2002. Genetic characterization of infectious hematopoietic necrosis virus of coastal salmonid stocks in Washington State. J. Aquat. Anim. Health 14:25-34.
Emmenegger EJ, KentopfE, Thompson TM, Ranson J, Pittam S, Ryan A, Keon D, Carlino J, Troyer RM, Garver KA, Life R, Kurath G. Development of an aquatic pathogen database (AquaPathogenX) and its utilization in tracking emerging fish virus pathogens in North America. in preparation.
Garver KA, Troyer RM, Kurath G. 2003. Two distinct phylogenetic clades of IHN virus overlap within the Columbia River basin. Dis. Aq. Org. 55:187-203.
Jonstrup SP, Schuetze H, Kurath G, Gray T, Jensen BB, Oleson NJ. 2010. An isolate and sequence database of infectious hematopoietic necrosis virus (IHNV). J. Fish Disease 33:469-471.
Kurath G, Garver KA, Troyer RM, Emmenegger EJ, Einer-Jensen K, Anderson ED. 2003. Phylogeography of infectious hematopoietic necrosis virus in North America. J. Gen. Virol. 84:803-814. DOI 10.1099/vir.0.18771-0
Rudakova SL, Kurath G, Bochkova EV. 2007. Occurrence and genetic typing of infectious hematopoietic necrosis virus in Kamchatka Russia. Dis. Aquat. Org. 75:1-11.
Troyer RM, LaPatra S, Kurath G. 2000. Genetic analyses reveal unusually high diversity of infectious hematopoietic necrosis virus in rainbow trout aquaculture. J. Gen. Virol. 81:2823-2832.
Troyer RM, Kurath G. 2003. Molecular epidemiology of infectious hematopoietic necrosis virus reveals complex virus traffic and evolution within southern Idaho aquaculture. Dis. Aquat. Org. 55:175-185.